A group from the University Medical Center Utrecht in the Netherlands has shown that new gene editing technology can be used to impair viral replication and clear latent herpesvirus infections. The group used a CRISPR-Cas system to target viral genetic elements that completely eliminated CMV and HSV1 replication. They were also able to clear latent EBV from transformed human tumor cells.
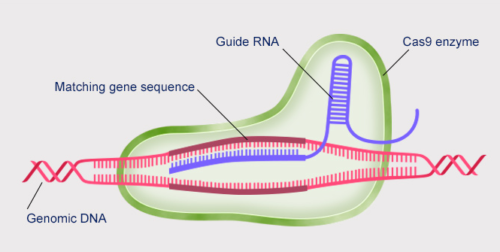
CRISPR comprises a single molecule of RNA that binds to the target gene (dark red) and delivers a DNA cutting enzyme (in this case Cas9) to the site.
CRISPR (clustered regularly interspaced palindromic repeats) Cas (CRISPR associated) is revolutionary technology that won the “Science Magazine Breakthrough of the Year” award in 2012 and 2013, and was named as one of the top 10 “breakthrough” technologies by MIT in 2014 and 2016.
The CRISPR system takes advantage of the adaptive defense mechanisms used by prokaryotes to combat the introduction of foreign DNA. Upon cell entry, foreign DNA particles are integrated into CRISPR sequences as spacers. The new genomic sequences are transcribed, resulting in the production of CRISPR RNAs (crRNAs), which can then target matching foreign DNA and, with associated Cas proteins, cause degradation of the DNA. In recent years, this system has been modified to target genes in human cells, and it has been successful in modifying RNA viruses such as HIV.
In the new study, the investigators designed guide RNAs (gRNAs) that targeted genes in CMV, HSV-1, and EBV and introduced double stranded DNA breaks. DNA cleaved by CRISPR is subsequently repaired by the cell’s own machinery, often resulting in insertions and deletions that may disrupt the function of the target protein.
Using recombinant EBV expressing eGFP to visualize the presence of the latent virus, the investigators found that gRNAs targeting EBV nuclear antigen 1 (EBNA1) and sequences in the viral origin of replication (OriP) caused a marked loss of EBV genomes from cells. Combinations of two different gRNAs increased the effectiveness of the strategy; qPCR revealed that the sequential application of two vectors reduced EBV genome content in the cells by over 95%.
To investigate the effects of CRISPR on CMV, genes involved in CMV replication were targeted, and 4 gRNAs were designed per gene. As in the EBV targeting, a lentiviral delivery system was used to introduce the gRNAs, and the resulting alterations at target sites on essential genes effectively inhibited CMV replication.
Active replication was also targeted in HSV-1, and most of the gRNAs for essential genes effectively reduced replication, but unlike those for CMV, some gRNAs aimed at nonessential HSV-1 genes also reduced replication. As in the EBV experiments, a combination of two gRNAs were most effective, resulting in the complete loss of infectious particles. However, targeting of the HSV-1 genome in the latent phase was ineffective.
In addition to a reduction in functional protein due to mutations arising from improper repair at the CRISPR cleavage site, the authors suggest that the cuts may also disrupt the generation and packaging of viral particles, and the increased effectiveness observed when using two gRNAs may be a result of genome fragmentation. They note that some mutations stemming from gRNA activity may cause mutations that do not reduce viral fitness. By using multiple gRNAs, however, the amount of active mutants can be lowered as the viral genome is fragmented. No off-target editing was observed during these experiments, but this is an important area of concern in a clinical setting, as cleavage at a site in the human genome could be harmful.
Current antiviral therapies for beta-herpesvirus infections (ganciclovir, cidofovir and foscarnet) target the viral DNA polymerase during active infection, but do nothing about the latent viral activity. HHV-6A and B latent infections can result in potent cytokine and chemokine activity in the absence of replication.
The effectiveness of certain gRNAs in reducing herpesvirus replication and genome content suggests a potential for CRISPR as a possible treatment strategy targeting latent and active HHV-6 at some point in the future.
For more information on current treatment options for HHV-6, visit our HHV-6 Treatment page.
Read the full paper here: CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections.

