iciHHV-6B, when integrated into chromosome 22q, was strongly associated with SLE and with SLE severity.
About 1% of humans inherit the HHV-6A or HHV-6B genome. Whether this inheritance renders people vulnerable to (or protected from) any human diseases is an important unanswered question.
A multi-institutional team from Japan, led by investigators at Riken, a national scientific research institute, analyzed whole-genome sequencing datasets from 6,321 Japanese individuals. They evaluated whether people with various autoimmune diseases—SLE, psoriasis, rheumatoid arthritis, multiple sclerosis, or pulmonary alveolar proteinosis (PAP)—were more likely to have iciHHV-6A/B than people without these diseases.
In a report published in Nature Genetics, the team found that iciHHV-6B, but not iciHHV-6A, was found significantly more often in people with SLE and PAP. After multivariable adjustment for potential confounders (e.g., age and gender), the association of iciHHV-6B and SLE remained highly statistically significant: the adjusted odds ratios and P-values for SLE and PSP were 6.9, P=0.006 and 7.0, P=0.007, respectively. Although there were several integration sites for iciHHV-6B in the population studied, only integration of the viral genome in chromosome 22q was significantly associated with SLE. In all four people with iciHHV-6B and SLE, the full-length viral genome was integrated. However, there was no detectable expression of HHV-6B genes in any of the cell types studied.
To validate the associations they had found in a Japanese cohort, the team then examined data from 121,494 people of European ancestry and 243,247 people of multiple ancestries in the NIH’s All of Us database. The adjusted odds ratios for SLE in the two populations were 2.7 and 2.4, respectively. However, the association of iciHHV-6B with PAP was not replicated in the All of Us cohort. As with the Japanese cohort, there was no association of iciHHV-6A and the autoimmune diseases studied in the All of Us cohort.
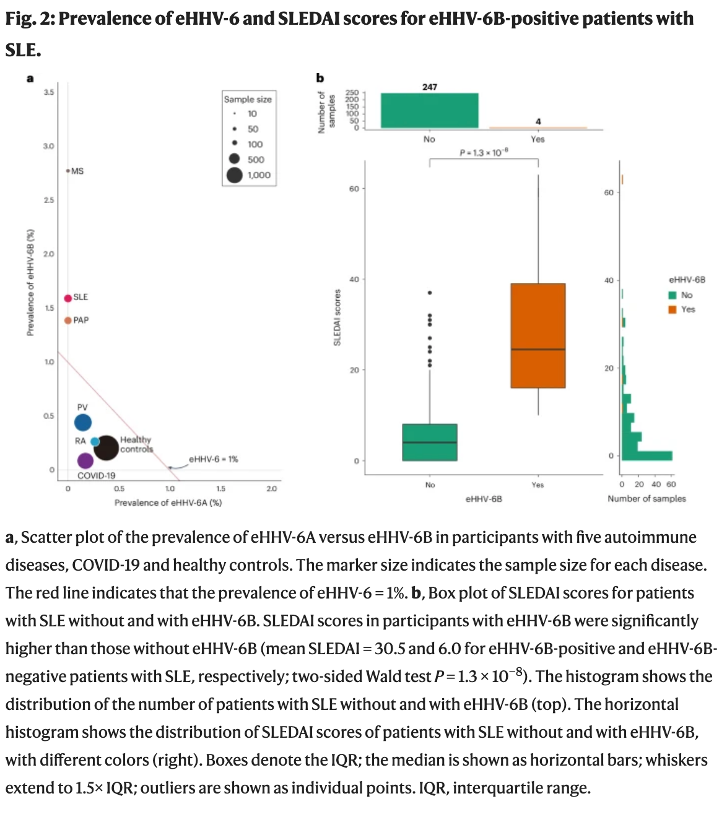
Consistent with iciHHV-6B having a causal role in the development of SLE, and its severity, the team found that the clinical severity score among people with SLE who also had iciHHV-6B was much higher than among people with SLE who did not have iciHHV-6B (see figure, below).
In addition, distinctive immune responses were detected against HHV-6B in patients with SLE. Specifically, responses against one specific region of one HHV-6 peptide—amino acids 476–490) in the immediate-early A (IE-A) transactivator—were significantly higher among SLE patients with iciHHV-6B than in those without iciHHV-6B. This is consistent with a previous study from the University of Washington that found that spontaneous gene expression in iciHHV6 individuals leads to increased HHV-6 IE-1 and CMV antibody response, with the IE-1A transactivator and U100 showing the highest level of expression (Peddu 2019).
In addition, expression of antiviral genes (particularly interferon-stimulated genes) was significantly more frequent in monocytes and dendritic cells of the four cases of SLE who had iciHHV-6B than in healthy controls.
This study also found that people with SLE had higher viral loads of anelloviruses in people with SLE, but no validation of this finding in another cohort was reported.
Several previous reports have linked iciHHV-6 to diseases. Most of these studies have been underpowered to find real disease associations. This study found a robust association of iciHHV-6B and SLE in both a Japanese and European population. It remains possible that it is not the integrated viral genome that is responsible for the increased risk of SLE; instead, other genetic loci that are tightly linked to the integrated viral genome may be the loci responsible for the increased risk of SLE.
Read the full text: Sasa 2024

