Outcomes worse for the subtype linked to EBV and HHV-6.
Classical Hodgkin’s lymphoma (HL), a B cell lymphoma, often is cured by first-line treatment. However, some patients develop relapsed disease and then succumb. The different disease trajectories have suggested that there may be biologically different subtypes of the disease, and that identifying these subtypes might help focus treatment.
It has been challenging to genomically characterize HL because biopsy specimens contain relatively few malignant cells. However, studies of circulating tumor DNA (ctDNA)—so-called “liquid biopsies”—have overcome this limitation.
A multi-institutional German team reports in the Journal of Clinical Oncology that it used ctDNA to identify three subsets of HL, and then confirmed the designation of these subsets in a separate validation sample of patients. The initial group included 243 people with HL and the validation group included 96 people. In both groups, the three subsets that emerged—and the features that characterized each subset—are as follows:
Inflammatory-immune escape HL –
- Somatic copy number variations in 12q12-12q13.3, 9p24.1, 2p16.1, 4q34.3-4q35.1, and6q23.3 were the defining genomic features
- Mutational burden increased, compared to the Virally-driven HL subset
- Increased numbers of B cells, TFH cells and TREG cells, compared to other subsets
- Decreased numbers of cytotoxic CD8+ cells, dendritic cells, eosinophils, γδ cells, mast cells, NK cells, compared to other subsets
- Fever, night sweats and weight loss (“B symptoms”) more common
Virally-driven HL
- Significant enrichment was noted for EBV and, to a somewhat lesser degree, with HHV6 DNA—but not for other viruses
- The association with EBV was validated with histopathologic confirmation: 21/22 cases under this subtype stained positive for EBV
- Increased numbers of cytotoxic CD8+ cells, dendritic cells, eosinophils, γδ cells, mast cells, NK cells, compared to other subsets
- Fever, night sweats and weight loss (“B symptoms”) more common
- Mutational burden much decreased, compared to the other two subsets
- Copy number variations found were predicted to affect NFκB signaling, MHC-1 presentation, interferon signaling, and cell cycle regulation
Oncogene-driven HL
- Greatly increased mutational burden, particularly involving these genes: TNFAIP3, ITPKB, IGLL5, and B2M.
- Somatic copy number variations in 2p16.1, 4q24.3-4q35.1, and 6q23.3
- B symptoms less common
The differences between the three subsets of HL are illustrated in Figure 1, below:
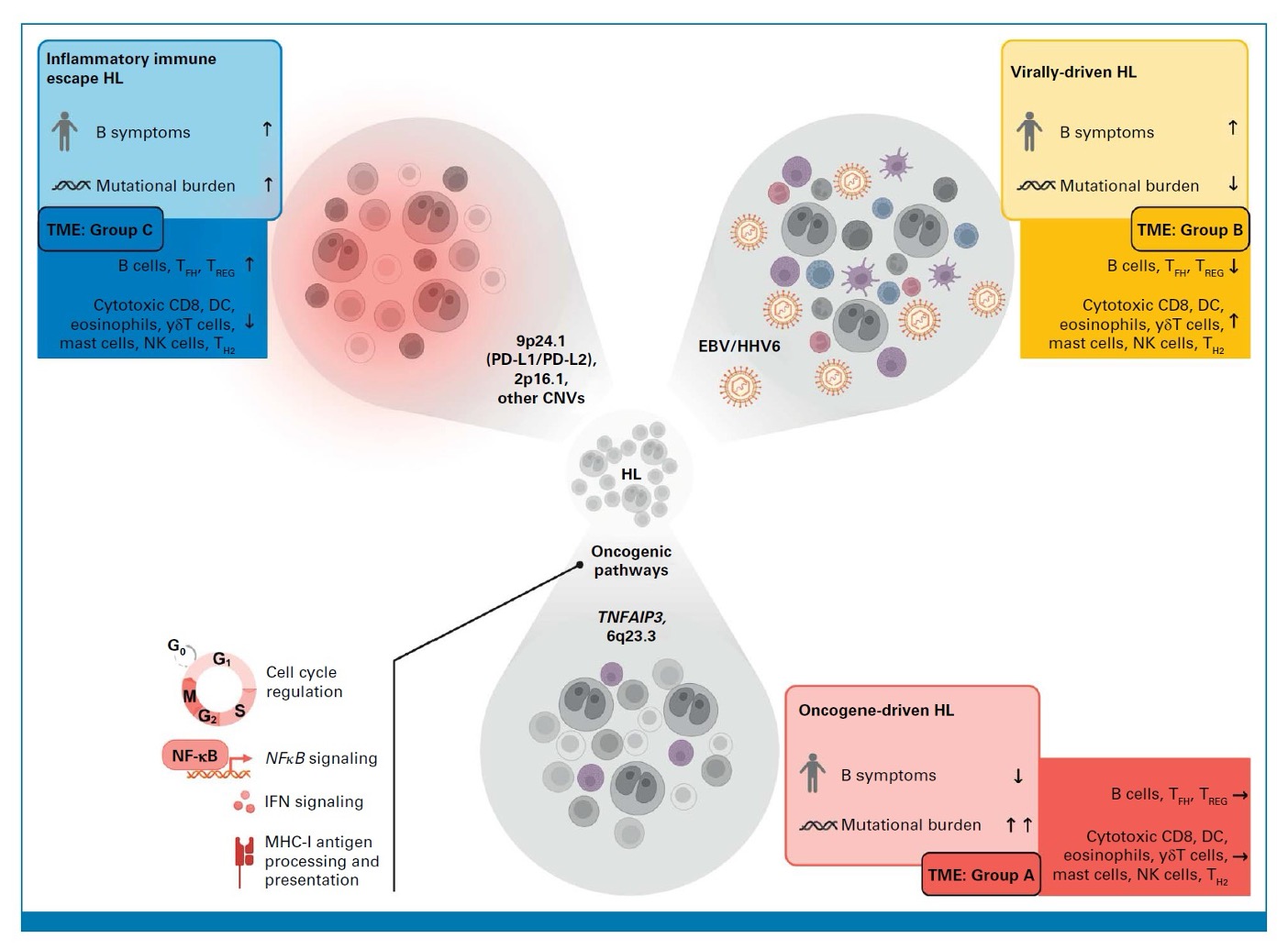
Figure 1. Schematic illustration describing key features of inflammatory immune escape HL, virally-driven HL, and oncogene-driven HL. The tumor microenvironment (TME) groups were characterized by gene expression profiles: TME Group A lacked overexpression of immune-associated genes; TME Group B had increased expression of several genes that indicated an inflammatory response by myeloid cells, and an upregulation of genes involved in viral infection. Patients in the virally-driven HL group were most likely to be in this subset; The TME Group C was characterized by upregulation of a different set of genes, including BCL2.
Prognosis was different in the three subtypes. Progression-free survival was longest in the oncogene-driven subtype, shorter in the virally-driven subtype, and shortest in the inflammatory-immune escape subtype. Whether further study of these subtypes will also reveal preferred treatment approaches, and therefore improved prognosis, for each subtype remains to be determined.
HHV-6 has long been suspected to play a role in Hodgkin’s lymphoma. In 2010, French investigators demonstrated the presence of HHV-6B in 40% of the nodular sclerosis subset of HL, and found an HHV-6B specific protein DR-7 in 74% of HL tissues (Lacroix 2010). A Yale pathologist found that HHV-6B was localized in Reed-Sternberg cells in the nodular sclerosis subset of Hodgkins lymphoma (Siddon 2012). A review on HHV-6 and malignancy published in 2018 concluded that “HHV-6, if not directly oncogenic, may act as a contributory factor that indirectly enhances tumor cell growth, in some cases by cooperation with other viruses” (Eliassen 2018).
Read the full text: Heger 2024