A team at University of Pittsburgh analyzed a large database of deep sequencing data from tumor and control tissues to look for viral sequences in 22 different cancers. They were surprised to find several herpesviruses in gastrointestinal cancers but not in control tissues.
The investigators utilized the Cancer Genome Atlas (TCGA), which was designed to enable discovery of pathogens in the tissues of cancer patients. HHV-6, CMV, and EBV were the most commonly detected viruses in stomach and colorectal cancers. HHV-6 was identified in 3.9% of stomach cancer and 4.7% of colon cancer samples, but was found in none of the normal stomach or colon controls. As expected, HPV was found in cervical cancer and hepatitis B and C in liver cancer.
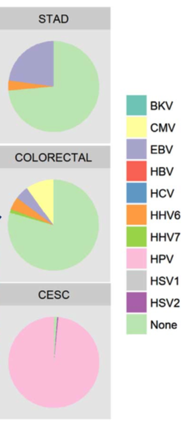
Viral sequences found in stomach adenocarcinomas, colorectal and cervical squamous cell carcinomas. Source: Virology 2018
The herpesviruses were present at a frequency (21% of rectal, 18% of colon, and 18% of stomach tumors) comparable to that of human papillomavirus in head and neck squamous cell carcinomas (20.1%). In some cases, HHV-6 was detected by both DNA-sequencing and RNA-sequencing, signaling that the positivity was not a result of contamination. While the association between HHV-6 and other cancers was not as striking, the virus was present more often in pancreatic and head and neck squamous cell carcinomas compared to matched controls (5% vs 0% and 2.1% vs 1.2%, respectively).
Gastrointestinal cancer (GIC) begins as a benign polyp and progresses into a carcinoma. In one study, HHV-6 antigen was detected in adenomatous polyps but not in healthy mucosal tissues (Halme 2013), and reactivation in the GI tract of immunosuppressed individuals, as well as in some immunocompetent individuals, results in diarrhea, colitis, and bacterial infection of the digestive tract (Halme 2008, Revest 2011).
An earlier study by Cao et al. also used the sequences provided by the CGA and found HHV-6 at a high prevalence in GICs (Cao 2016). Specifically, HHV-6 was detected in 26 cases of colon adenocarcinomas (5.8% of total cases), 9 cases of stomach adenocarcinomas (3.5%), 7 cases of esophageal cancer (6.4%), and 6 cases of rectal adenocarcinomas (3.7%). In contrast to the present study, the former work did not utilize both RNA sequencing and DNA sequencing, and the prevalence of viruses was not determined among the controls for many cancers.
As is often the case, it is difficult to know what the exact role of HHV-6 is in the setting of GIC. In samples that were positive for HHV6, the team could not determine whether the expression pattern was indicative of latent or lytic infection because of the low coverage of most genes in the viral genome. Moreover, it was not possible to know which cells harbored the virus. The strongest support for a causative role for HHV-6 in GIC would result from finding HHV-6 in the tumor cells themselves, instead of in infiltrating lymphocytes.
Read the full paper here: Cantalupo 2017.