Noam Stern-Ginossar and her team have posted their unbiased atlas of the HHV-6 proteome on bioRxiv. They describe hundreds of new reading frames and three highly abundant HHV-6 encoded long non-coding RNAs.
Traditional genome profiling has been based on open-reading frame analysis. More recent techniques include RNA sequencing, genome-wide translation mapping, and others, which have led to the discovery of hundreds of new viral open reading frames and the re-annotation of parts of the viral genome.
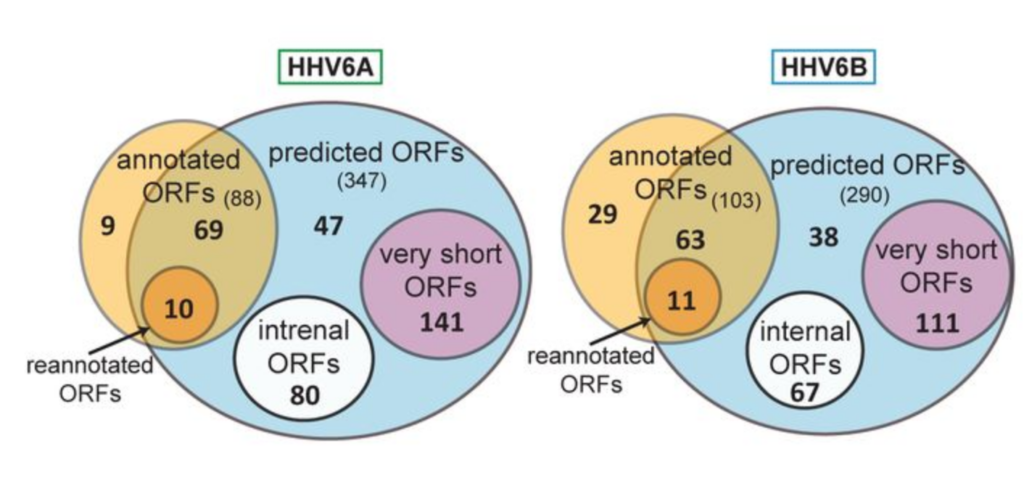
The investigators from the Weizmann Institute of Science and from the Hebrew University Hadassah Medical School used ribosome profiling and RNA sequencing to further understand the HHV-6 genome. This study resulted in more accurate mapping of both known and novel genomic features ultimately leading to a full inventory of HHV-6 translation products. Using RNA-sequencing the investigators were able to find 24 of the 26 previously discovered splice junctions in HHV-6A and 37 novel ones. Similarly, they found all 24 of the previously discovered HHV-6B splice junctions and 44 novel ones. Among these, dozens were conserved between HHV-6A and HHV-6B. Investigators also identified hundreds of novel open reading frames (ORFs) in both HHV-6A and HHV-6B and three sections of long non-coding RNA (lncRNA) that are conserved between HHV-6A and HHV-6B. Furthermore, by comparing ribosome profiling data with RNA-sequencing data the investigators were able to determine which open reading frames were likely to have translational products.
This in-depth study of the HHV-6A and HHV-6B genomes combined with the results from their previous study of the human cytomegalovirus (HCMV) genome (Stern-Ginossaret al., 2012) allowed the investigators to determine sections of genomic and functional conservation among the betaherpesvirus family. They found that several of the upstream ORFs and internal ORFs found in HHV-6 are also found in HCMV. Additionally, one of the lncRNAs found in HHV-6 shares similarities in function to a lncRNA found in HCMV.
Read the unpublished draft on bioRxiv: Finkel 2019