 A group led by Yasuko Mori in Japan has analyzed the crystal structure of HHV-6B U14, an important accomplishment for the understanding of HHV-6. Human herpesvirus 6B encodes numerous tegument proteins that make up the viral matrix. One of these tegument proteins is U14. In addition to being necessary for viral propagation, it is able to regulate host cell responses by interacting with host factors such as tumor suppressor p53.
A group led by Yasuko Mori in Japan has analyzed the crystal structure of HHV-6B U14, an important accomplishment for the understanding of HHV-6. Human herpesvirus 6B encodes numerous tegument proteins that make up the viral matrix. One of these tegument proteins is U14. In addition to being necessary for viral propagation, it is able to regulate host cell responses by interacting with host factors such as tumor suppressor p53.
The virions of herpesviruses share a common structure. In addition to the capsid, there is an envelope. Between these two is a pool of tegument proteins, which not only act as structural components but also serve other important functions.
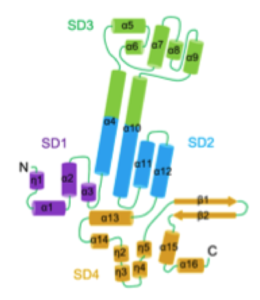
The researchers constructed U14-NTD in E. coli with MBP, which corresponds to the N-terminal region since the C-terminal region was unstable. U14-NTD was found to have an elongated helix-rich structure composed of sixteen a helices, four 310 helices, and two b strands. The researchers divided the structure into four subdomains (SDs) (see figure). In SD4, the two b strands protrude and form a b hairpin.
In the crystal structure, U14-NTD forms a dimer, where the long axes of the monomers run in an antiparallel orientation and the two b hairpins cross each other. All four SDs are involved in the dimer interface. Interestingly, there is an internal cavity within the dimer interface. On each monomer is a negatively-charged surface between SD2 and SD4 and a positively-charged surface between SD2 and SD3. Upon dimerization, the negatively-charged area of each monomer faces the positively-charged area of the other monomer. Surrounding these areas are a total of 40 hydrogen bonds, indicating tight and specific dimerization. The dimerization of U14 suggests that this protein acts as a scaffold in the viral matrix.
Along the outside surface of the U14-NTD dimer, there are multiple clusters of positive and negative electrostatic potential. The b hairpin of each monomer contains six negatively-charged residues and the region 342-378 of SD4 contains 11 negatively-charged residues. On the opposite side of the monomer, there are seven positively-charged residues, as well as ten negatively-charged residues.
On the b hairpin, there are three amino acids, L424, E425, and V426, which caused a defect in viral growth when they were deleted or substituted with alanines. As L424 and V426 are part of a hydrophobic patch that is solvent-exposed, the researchers believe that the b hairpin may be a key functional site of HHV-6 U14. In a previous study, deletion of the amino acids corresponding to L424, E425, and V426 resulted in a lack of interaction with tumor suppressor p53 (Mori 2015). Additionally, the interaction between U14 and another viral protein, U11, was abolished when the corresponding residues of L424, E425, and V426 were deleted.
The group performed multiple sequence alignment for HHV-6B U14, HHV-6A U14, HHV-7 U14, and HCMV UL35. It was found that U14-NTD was a core part conserved among all members. HHV-6A U14 and HHV-7 U14 were well aligned with HHV-6B U14 across all SDs while HCMV UL35 was well aligned except for SD4. It was also found that HHV-6A U14 and HHV-7 U14 had almost identical residues to HHV-6B U14 at the dimer interaction sites, indicating that these proteins also form a dimer. However, the residues were different for HCMV UL25 and UL35, suggesting that dimerization appears to be specific to HHV-6 and HHV-7.
For more information, read the full paper, Wang 2016.

