Investigators at Mt Sinai used “big data” models to determine that the genes involved with fighting Alzheimer’s are the same ones that fight viruses. They found HHV-6A and HHV-7 to be more abundant in Alzheimer’s brains, and singled out HHV6-A as a key modulator of the genes involved in amyloidosis and neuronal death.
Both HHV-6A and HHV-7 DNA viral loads and transcription were increased among AD patients, and this was observed across multiple brain regions and multiple cohorts but not in healthy aging controls. Host genes affected by HHV-6A were all associated with Alzheimer’s traits and risk genes. Also HHV-6A correlated strongly with dementia scores, neuronal death and intensity of amyloid plaque.

Joel Dudley, PhD – Associate Professor of Genetics and Director, Next Generation Healthcare Institute, Mt Sinai, New York
The group, led by Joel Dudley at the Icahn School of Medicine at Mt. Sinai, began by mapping and comparing differences among gene regulatory networks in preclinical AD cases, in order to explore early changes during AD development, and found that C2H2 zinc finger transcription factor binding motifs and G-quadruplex sequences, which are associated with viral biology, were differentially regulated between AD and control brains. In light of this finding, the investigators examined the AD-associated virome, finding that genes from several viral species, but particularly HHV-6A and HHV-7, were abundantly expressed in patient brains. HSV-1 latency transcripts were also more abundant in AD brains.
Furthermore, consistently increased levels of HHV-6A and HHV-7 RNA in patient brains was observed in multiple cohorts and several brain regions. In contrast, heightened expression of HHV-6A and HHV-7 genes was not seen among brain tissues from individuals with pathological aging, indicating that the pattern of viral activity characteristic of AD is not a ubiquitous feature of neurodegenerative disease as a whole. On the other hand, higher expression of HHV-6A was found among progressive supranuclear palsy specimens, suggesting that HHV-6A may contribute to other neurological conditions.
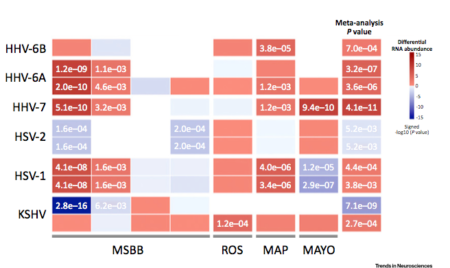
Differential abundance of viral transcripts in AD vs control brains
Source: Readhead 2018, Neuron
Dudley’s team determined that many host DNA markers were significantly associated with high levels of virus, and several viral genes (including the HHV-6A unique region and the HHV-7 DR-1 gene) were significantly associated with AD-associated genes, including PSEN1, BACE1, APBB2, CLU, BIN1, and PICALM. HHV-6A also appeared to interact with several transcription factor networks that have links to AD risk. The group also determined that several viruses affected expression of nucleotide regulating proteins, indicating that dysregulation of protein translation may be involved in AD progression.
The team also performed causal inference analysis that indicated that HHV-6A contributed to neuronal loss. In fact, HHV-6A was the only virus associated with cell fraction changes in several brain regions. In addition, genes that were regulated by HHV-6A and were involved in neuronal loss overlapped with risk loci for AD. One such gene encodes miR-155, which was found to be suppressed by HHV-6A. Moreover, the offspring of miR-155-knockout mice and APP/PS1 amyloidosis strain mice exhibited larger, more frequent cortical amyloid plaques and higher levels of amyloid-beta conformers than mice with normal miR-155 levels, and in the knockout mice, the differentially expressed host genes overlapped with those that were upregulated by HHV-6A.
MiR-155 has been linked to aspects of malignancy, innate immunity, and autoimmunity (Bernecker 2012). HHV-6A-induced suppression of this microRNA was similarly found by Caselli et al. and Rizzo et al., both in natural killer cells (innate immune cells) (Rizzo 2017) and in thyroid cells (Caselli 2017) in vitro.
Host genes upregulated by HHV-6A are enriched for a heterogeneous set of AD risk and biomarker-associated genes. Shown here are the HHV-6A/AD-associated gene subnetwork and detected interactions with additional viruses. Source: Neuron, 2018[/caption]
The results of this study highlight the potential for HHV-6 to produce sub-clinical, tissue-specific changes that produce serious pathology over time as a result of long-term persistent infection. It has been suggested that this type of persistent infection may contribute to a variety of illnesses, including lymphoproliferative disorders and cancer (Eliassen 2018). Persistent HHV-6A infection of the uterus, for example, appears to affect immune cell populations and immune mediators, thereby leading to infertility (Marci 2016, Caselli 2017, Coulam 2018).
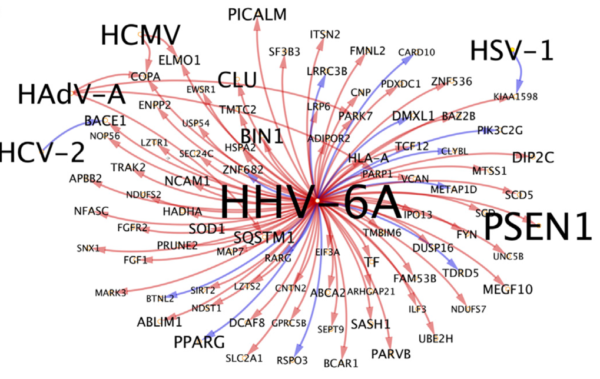
Host genes upregulated by HHV-6A are enriched for a heterogeneous set of AD risk and biomarker-associated genes. Shown here are the HHV-6A/AD-associated gene subnetwork and detected interactions with additional viruses. Source: Neuron, 2018
In a mouse model, HHV-6A but not HHV-6B established persistent infection in the brain and was associated with lymphocytic infiltration and upregulation of CCL5 (Reynaud 2014). In vitro infection of glial cells with HHV-6A resulted in upregulation of certain proinflammatory chemokines, including CCL2, CCL5, and CXCL10, which appeared to rely on toll-receptor 9 signaling. No clinical signs of disease were noted in the mice during the course of observation, which lasted up to nine months. Presumably, altered cellular functioning over a period of many years could result in neuropathology and clinical manifestations.
As is often the case with HHV-6, it is difficult to definitively determine whether higher viral loads of HHV-6 contribute to the disease or whether the elevated viral load is a result of the modified immune environment resulting from the disease. The presence of HHV-6A RNA allows for differentiation between latent and active virus; in this case, the virus present in AD brains was transcriptionally active and less likely to be an insignificant bystander.
The authors note that evaluation of the development and progression of AD phenotypes in chronic HHV-6A-infected animal models could provide further evidence in favor of a causative role for the virus in AD.
Find the full text: Readhead 2018.
HHV-6A and HHV-6B have been investigated in several neurological disorders, including encephalitis/encephalopathy, multiple sclerosis, chronic fatigue syndrome, post-transplant Parkinsonism, and post-transplant delirium. In 2014, HHV-6 (untyped) DNA was found in 17% of brain tissues from patients with Alzheimer’s disease (AD), and HHV-6 was found more frequently in the blood of AD patients than in healthy controls (23% vs 4%, respectively) (Carbone 2014). The same group went on to find genotypic variants that correlated with both AD development and HHV-6 DNA positivity (Licastro 2015). A third study documented lower HHV-6 IgG reactivity among patients with AD compared to healthy controls, possibly suggestive of impaired immunity against HHV-6 leading to increased reactivation in the brain (Westman 2017).
