Next-generation sequencing proves valuable for identifying CNS infections in the immunocompetent.
The diagnosis of suspected central nervous system (CNS) infection has long been challenging: multiple microorganisms can cause CNS infections, and traditional diagnostic methods for identifying each of them can be different.
For the past decade, next-generation (NGS) sequencing of cerebrospinal fluid (CSF) has offered the promise of a single technique that could identify multiple different types of pathogens. A team from the University of California San Francisco (UCSF) that has pioneered NGS of CSF now reports its experience over the past 7 years.
The team tested 4,828 CSF samples with NGS as well as with traditional methods. The samples came from around the country. For a group of over 1,164 samples from 1,053 patients who received medical care at UCSF, the team also assessed the role of other clinical information besides CSF testing. Of this latter group, 64% were immunocompetent but the remainder were immunocompromised in some way.
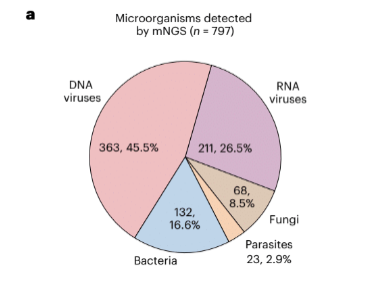
In the larger group of samples from around the country, after excluding microbes judged to be contaminants, likely responsible pathogens were identified in 14.4% of CSF samples. The identified pathogens were predominantly viruses (72%), but also bacteria (16.6%), fungi (8.5%) and parasites (2.9%).
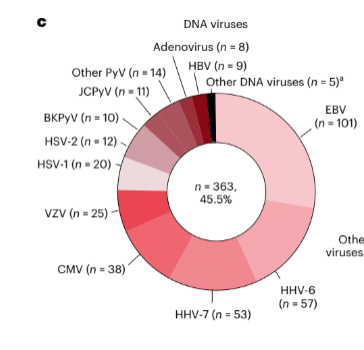
Among the viruses, the most frequently identified were EBV (101 cases), HIV (92 cases), HHV-6 (57 cases), HHV-7 (53 cases), CMV (38 cases), West Nile Virus (28 cases), VZV (25 cases), HSV-1 (20 cases), HSV-2 (12 cases). Thus, HHV-6 and HHV-7 were among the most frequently identified viruses. In the smaller group of over a thousand UCSF patients, a somewhat larger fraction of CSF samples contained likely pathogens identified by NGS: 21.8%.
The sensitivity, specificity and accuracy of mNGS testing for CNS infections were 63.1%, 99.6% and 92.9%, respectively. mNGS testing exhibited higher sensitivity (63.1%) than indirect serologic testing (28.8%) and direct detection testing from both CSF (45.9%) and non-CSF (15.0%) samples (P < 0.001 for all three comparisons). When only considering diagnoses made by CSF direct detection testing, the sensitivity of mNGS testing increased to 86%. The frequency of different types of pathogens, and the frequency of specific viruses within the larger group of DNA viruses, are shown in the figure below.
NGS testing was the most sensitive test for the detection of pathogens in people with clinically severe and diagnostically challenging CNS infections. NGS testing was the only or the first test to make the diagnosis in 67 (30.4%) of 220 infections. At the same time, the overall sensitivity of 63.1% for mNGS testing (86% in cases diagnosed by CSF direct detection methods, including mNGS) was not sufficient to replace conventional microbiologic testing. And NGS testing at UCSF, took longer to produce results (9 days, on average) than more traditional methods.
The high level of EBV found was suprising and may reflect the fact that 64% of the patients tested were immunocompetent. By contrast, in immunocompromised transplant patients tested for encephalitis, HHV-6 was by far the most common virus found, followed by EBV and CMV; in cord blood transplant patients, HHV-6 causes 95.7% of encephalitis cases (Toomey 2023).
The primary purpose of this report was to assess the value of NGS of CSF in diagnosing the cause of suspected CNS infections. The authors identify four virtues of NGS testing of CSF in such infections: 1) detection of unculturable, difficult-to-diagnose organisms, 2) broadly diagnosing viral infections, 3) identifying rare, unexpected infections and 4) aiding in public health investigations of outbreaks.
We cover this report because it also highlights how frequent both HHV-6 and HHV-7 may be as responsible pathogens in CNS infections. Many past studies of suspected CNS infections—particularly in immunocompetent people—have not even looked for these viruses, whether using traditional or newer molecular techniques.
Read the full text: Benoit 2024

