Short non-coding RNA’s provide cell-free, activity-specific viral signatures
Researchers from the Singapore-MIT Alliance for Research and Technology identified unique biomarkers to differentiate between live virus infection and dead/inert viral fragments in nucleic acid amplification tests (NAATs), demonstrating the potential for a novel and expeditious method of viral testing.
Traditional nucleic acid amplification tests (NAATs) lack the ability to distinguish between live/dead viruses or latent/reactivated states. Extracellular viral microRNAs (viral exmiRs)—short non-coding RNAs secreted by infected cells—can address this gap.
EBV miR-BART10-3p and HSV-1 miR-H5 were identified in spent media of infected T cells as able to distinguish between live infections from heat-inactivated or antibody-neutralized viruses. Additionally, detection was possible within 8 hours post-infection, with sensitivity down to 30 attomolar.
CMV miR-US5-2-5p and miR-US22-5p served as plasma biomarkers in latent HCMV-positive patients, who were negative for CMV by standard DNA testing.
A single donor was identified as having latent HHV-6 evidenced by detection of U94 gene. In this donor, HHV-6 miR-Ro6-4 was detected, but was absent in the other HHV-6 negative donors, demonstrating a possible specificity of the miR and a marker for efficient endogenous HHV-6 detection.
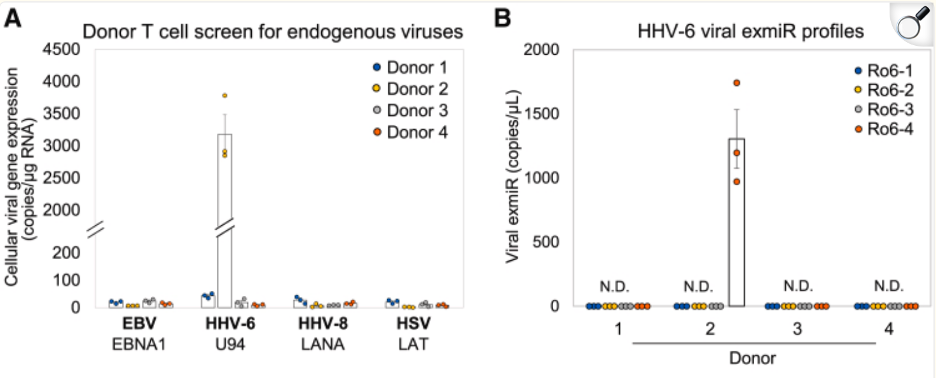
Four healthy human donor PBMC-derived T cell cultures screened for latency genes of common endogenous viruses in T cells using RT-dPCR: EBV-2 (EBNA1), HHV-6B (U94), HHV-8 (LANA), and HSV-1 (LAT). B) quantification of several HHV-6 exmiR using RT-qPCR.
A summary of markers of viral exmiR biomarkers of live adventitious, endogenous latent, and reactivated viruses found in this study can be found below:

Read the full text: Chan 2025