EBV was found in 28.6%; HHV-6 and CMV were found in 18.4% of cases.
Bloodstream infections (BSIs) are critical yet challenging to diagnose due to the limitations of conventional blood cultures (BCs), particularly when patients receive prior antimicrobial therapy or when pathogens are non-culturable
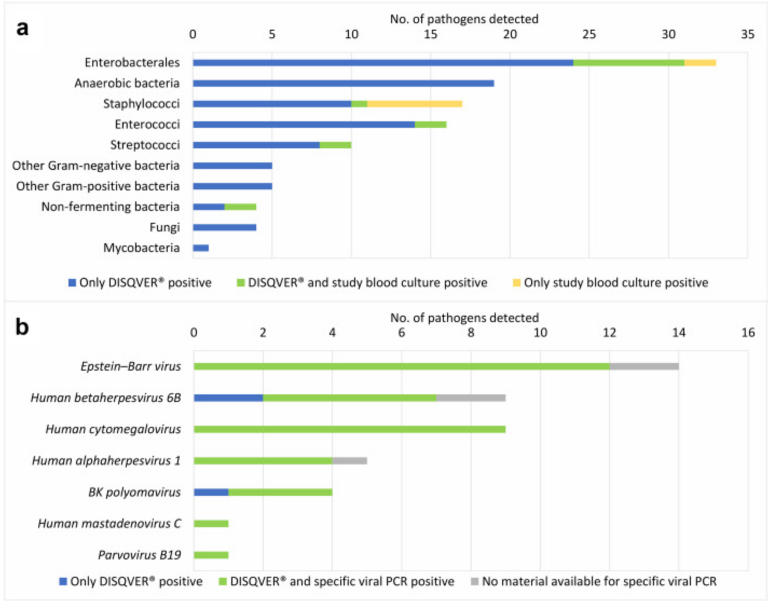
Investigators in Germany used metagenomic next generation sequencing (NGS) of cell-free DNA (cfDNA) in plasma followed by DISQVER©, a software algorithm based on a curated database of 16,000 microorganisms to study samples from patients suspected of bloodstream infections to study the performance of this unbiased approach to pathogen detection. The sequencing tool was developed by Noscendo GmbH, A Germany provider of software-based pathogen identification.
This retrospective study was conducted at University Hospital Erlangen, Germany with 190 samples analyzed from 147 patients, of which 67 patients were from the ICU.
The novel methodology led to a change in therapy in 20 (13.6%) patients, of which 10 were post-HSCT. Of these 20, viruses were detected in 6 patients, which led to the initiation of antiviral therapy or cessation of antibiotic therapy.
Read the full text: Esse 2025

